"""
Created on Mon Apr 15 19:43:04 2019
Updated on Wed Jan 29 10:18:09 2020
@author: created by Sowmya Myneni and updated by Dijiang Huang
"""'\nCreated on Mon Apr 15 19:43:04 2019\n\nUpdated on Wed Jan 29 10:18:09 2020\n\n@author: created by Sowmya Myneni and updated by Dijiang Huang\n'import matplotlib.pyplot as plt
import tensorflow as tf
print("Num GPUs Available:", len(tf.config.list_physical_devices('GPU')))Num GPUs Available: 1
import os
import multiprocessing
# Number of logical CPUs
num_cores = multiprocessing.cpu_count()
print(f"Number of CPU cores available: {num_cores}")Number of CPU cores available: 16
# Configure TensorFlow to use all available CPU cores
#tf.config.threading.set_intra_op_parallelism_threads(0) # Use all intra-operation threads
#tf.config.threading.set_inter_op_parallelism_threads(0) # Use all inter-operation threadsFNN Sample SA¶
########################################
# Part 1 - Data Pre-Processing
#######################################
# To load a dataset file in Python, you can use Pandas. Import pandas using the line below
import pandas as pd
# Import numpy to perform operations on the dataset
import numpy as npVariable Setup¶
# Variable Setup
# Available datasets: KDDTrain+.txt, KDDTest+.txt, etc. More read Data Set Introduction.html within the NSL-KDD dataset folder
# Type the training dataset file name in ''
#TrainingDataPath='NSL-KDD/'
#TrainingData='KDDTrain+_20Percent.txt'
# Batch Size
BatchSize=10
# Epohe Size
NumEpoch=10Import Training Dataset¶
# Import dataset.
# Dataset is given in TraningData variable You can replace it with the file
# path such as “C:\Users\...\dataset.csv’.
# The file can be a .txt as well.
# If the dataset file has header, then keep header=0 otherwise use header=none
# reference: https://www.shanelynn.ie/select-pandas-dataframe-rows-and-columns-using-iloc-loc-and-ix/
dataset_train = pd.read_csv('Training-a1-a3.csv', header=None)dataset_train.info()<class 'pandas.core.frame.DataFrame'>
RangeIndex: 113322 entries, 0 to 113321
Data columns (total 43 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 0 113322 non-null int64
1 1 113322 non-null object
2 2 113322 non-null object
3 3 113322 non-null object
4 4 113322 non-null int64
5 5 113322 non-null int64
6 6 113322 non-null int64
7 7 113322 non-null int64
8 8 113322 non-null int64
9 9 113322 non-null int64
10 10 113322 non-null int64
11 11 113322 non-null int64
12 12 113322 non-null int64
13 13 113322 non-null int64
14 14 113322 non-null int64
15 15 113322 non-null int64
16 16 113322 non-null int64
17 17 113322 non-null int64
18 18 113322 non-null int64
19 19 113322 non-null int64
20 20 113322 non-null int64
21 21 113322 non-null int64
22 22 113322 non-null int64
23 23 113322 non-null int64
24 24 113322 non-null float64
25 25 113322 non-null float64
26 26 113322 non-null float64
27 27 113322 non-null float64
28 28 113322 non-null float64
29 29 113322 non-null float64
30 30 113322 non-null float64
31 31 113322 non-null int64
32 32 113322 non-null int64
33 33 113322 non-null float64
34 34 113322 non-null float64
35 35 113322 non-null float64
36 36 113322 non-null float64
37 37 113322 non-null float64
38 38 113322 non-null float64
39 39 113322 non-null float64
40 40 113322 non-null float64
41 41 113322 non-null object
42 42 113322 non-null int64
dtypes: float64(15), int64(24), object(4)
memory usage: 37.2+ MB
dataset_train.describe()Loading...
# Identify the column that contains the attack types
# Assuming the column with attack types is named based on the sample data ("neptune", "normal", etc.)
attack_column = dataset_train.columns[-2] # Second last column seems to have attack types
# Calculate the frequency of each attack type
attack_distribution = dataset_train[attack_column].value_counts()
plt.figure(figsize=(10, 6))
attack_distribution.plot(kind='bar', logy=True)
plt.title('A1 and A3 Training Dataset Distribution of Attack Types')
plt.xlabel('Attack Type')
plt.ylabel('Frequency')
plt.xticks(rotation=45)
plt.tight_layout()
plt.show()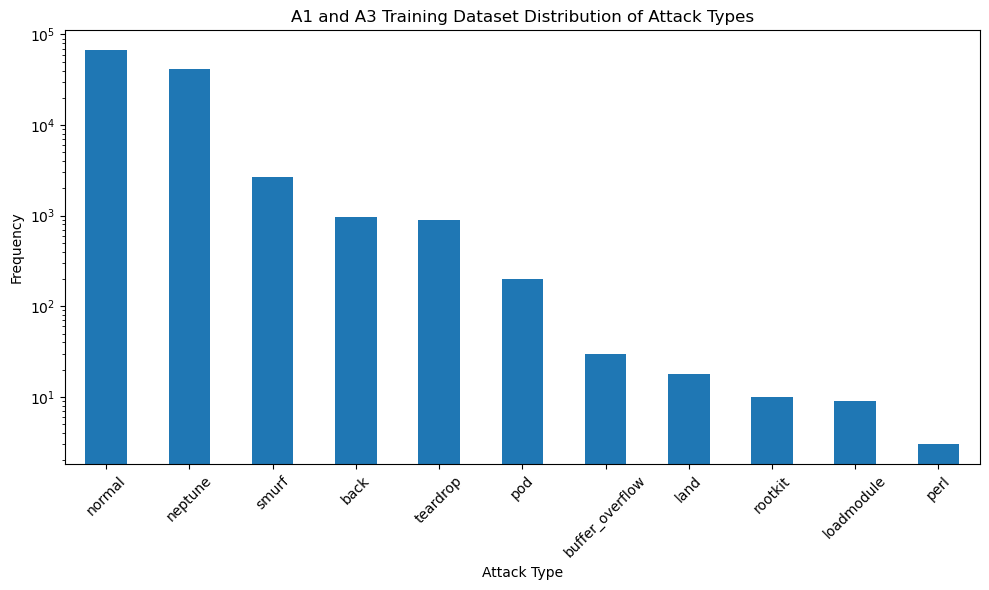
X_train = dataset_train.iloc[:, 0:-2].values
X_trainarray([[0, 'tcp', 'ftp_data', ..., 0.0, 0.05, 0.0],
[0, 'udp', 'other', ..., 0.0, 0.0, 0.0],
[0, 'tcp', 'private', ..., 1.0, 0.0, 0.0],
...,
[0, 'tcp', 'smtp', ..., 0.0, 0.01, 0.0],
[0, 'tcp', 'klogin', ..., 1.0, 0.0, 0.0],
[0, 'tcp', 'ftp_data', ..., 0.0, 0.0, 0.0]], dtype=object)X_train.shape(113322, 41)label_column_train = dataset_train.iloc[:, -2].values
label_column_trainarray(['normal', 'normal', 'neptune', ..., 'normal', 'neptune', 'normal'],
dtype=object)y_train = []
for i in range(len(label_column_train)):
if label_column_train[i] == 'normal':
y_train.append(0)
else:
y_train.append(1)
# Convert list to array
y_train = np.array(y_train)y_trainarray([0, 0, 1, ..., 0, 1, 0])y_train.shape(113322,)Import Testing Dataset¶
# Import dataset.
# Dataset is given in TraningData variable You can replace it with the file
# path such as “C:\Users\...\dataset.csv’.
# The file can be a .txt as well.
# If the dataset file has header, then keep header=0 otherwise use header=none
# reference: https://www.shanelynn.ie/select-pandas-dataframe-rows-and-columns-using-iloc-loc-and-ix/
dataset_test = pd.read_csv('Testing-a2-a4.csv', header=None)dataset_test.info()<class 'pandas.core.frame.DataFrame'>
RangeIndex: 15017 entries, 0 to 15016
Data columns (total 43 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 0 15017 non-null int64
1 1 15017 non-null object
2 2 15017 non-null object
3 3 15017 non-null object
4 4 15017 non-null int64
5 5 15017 non-null int64
6 6 15017 non-null int64
7 7 15017 non-null int64
8 8 15017 non-null int64
9 9 15017 non-null int64
10 10 15017 non-null int64
11 11 15017 non-null int64
12 12 15017 non-null int64
13 13 15017 non-null int64
14 14 15017 non-null int64
15 15 15017 non-null int64
16 16 15017 non-null int64
17 17 15017 non-null int64
18 18 15017 non-null int64
19 19 15017 non-null int64
20 20 15017 non-null int64
21 21 15017 non-null int64
22 22 15017 non-null int64
23 23 15017 non-null int64
24 24 15017 non-null float64
25 25 15017 non-null float64
26 26 15017 non-null float64
27 27 15017 non-null float64
28 28 15017 non-null float64
29 29 15017 non-null float64
30 30 15017 non-null float64
31 31 15017 non-null int64
32 32 15017 non-null int64
33 33 15017 non-null float64
34 34 15017 non-null float64
35 35 15017 non-null float64
36 36 15017 non-null float64
37 37 15017 non-null float64
38 38 15017 non-null float64
39 39 15017 non-null float64
40 40 15017 non-null float64
41 41 15017 non-null object
42 42 15017 non-null int64
dtypes: float64(15), int64(24), object(4)
memory usage: 4.9+ MB
dataset_test.describe()Loading...
# Identify the column that contains the attack types
# Assuming the column with attack types is named based on the sample data ("neptune", "normal", etc.)
attack_column = dataset_test.columns[-2] # Second last column seems to have attack types
# Calculate the frequency of each attack type
attack_distribution = dataset_test[attack_column].value_counts()
plt.figure(figsize=(10, 6))
attack_distribution.plot(kind='bar', logy=True)
plt.title('A2 and A4 Testing Dataset Distribution of Attack Types')
plt.xlabel('Attack Type')
plt.ylabel('Frequency')
plt.xticks(rotation=45)
plt.tight_layout()
plt.show()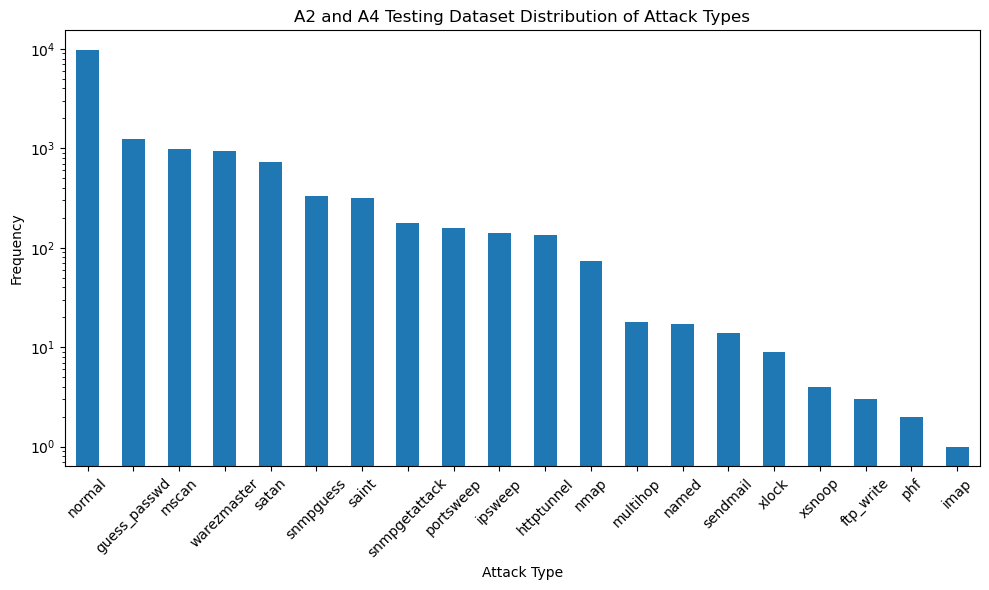
X_test = dataset_test.iloc[:, 0:-2].values
X_testarray([[2, 'tcp', 'ftp_data', ..., 0.0, 0.0, 0.0],
[0, 'icmp', 'eco_i', ..., 0.0, 0.0, 0.0],
[1, 'tcp', 'telnet', ..., 0.0, 0.83, 0.71],
...,
[0, 'tcp', 'http', ..., 0.0, 0.0, 0.0],
[0, 'udp', 'domain_u', ..., 0.0, 0.0, 0.0],
[0, 'tcp', 'sunrpc', ..., 0.0, 0.44, 1.0]], dtype=object)X_test.shape(15017, 41)label_column_test = dataset_test.iloc[:, -2].values
label_column_testarray(['normal', 'saint', 'mscan', ..., 'normal', 'normal', 'mscan'],
dtype=object)y_test = []
for i in range(len(label_column_test)):
if label_column_test[i] == 'normal':
y_test.append(0)
else:
y_test.append(1)
# Convert list to array
y_test = np.array(y_test)y_testarray([0, 1, 1, ..., 0, 0, 1])Encoding categorical data (convert letters/words in numbers)¶
# The following code work Python 3.7 or newer
from sklearn.preprocessing import OneHotEncoder
from sklearn.compose import ColumnTransformer
ct = ColumnTransformer(
# The column numbers to be transformed ([1, 2, 3] represents three columns to be transferred)
[('one_hot_encoder', OneHotEncoder(handle_unknown='ignore'), [1,2,3])],
# Leave the rest of the columns untouched
remainder='passthrough'
)
X_train = np.array(ct.fit_transform(X_train), dtype=np.float64)
X_test = np.array(ct.transform(X_test) , dtype=np.float64)X_trainarray([[0. , 1. , 0. , ..., 0. , 0.05, 0. ],
[0. , 0. , 1. , ..., 0. , 0. , 0. ],
[0. , 1. , 0. , ..., 1. , 0. , 0. ],
...,
[0. , 1. , 0. , ..., 0. , 0.01, 0. ],
[0. , 1. , 0. , ..., 1. , 0. , 0. ],
[0. , 1. , 0. , ..., 0. , 0. , 0. ]])X_train[0]array([0.00e+00, 1.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00,
0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00,
0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00,
0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 1.00e+00, 0.00e+00,
0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00,
0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00,
0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00,
0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00,
0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00,
0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00,
0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00,
0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00,
0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 1.00e+00, 0.00e+00,
0.00e+00, 4.91e+02, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00,
0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00,
0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00, 0.00e+00,
0.00e+00, 2.00e+00, 2.00e+00, 0.00e+00, 0.00e+00, 0.00e+00,
0.00e+00, 1.00e+00, 0.00e+00, 0.00e+00, 1.50e+02, 2.50e+01,
1.70e-01, 3.00e-02, 1.70e-01, 0.00e+00, 0.00e+00, 0.00e+00,
5.00e-02, 0.00e+00])X_train.shape(113322, 116)X_testarray([[0. , 1. , 0. , ..., 0. , 0. , 0. ],
[1. , 0. , 0. , ..., 0. , 0. , 0. ],
[0. , 1. , 0. , ..., 0. , 0.83, 0.71],
...,
[0. , 1. , 0. , ..., 0. , 0. , 0. ],
[0. , 0. , 1. , ..., 0. , 0. , 0. ],
[0. , 1. , 0. , ..., 0. , 0.44, 1. ]])X_test.shape(15017, 116)Perform feature scaling¶
# Perform feature scaling. For ANN you can use StandardScaler, for RNNs recommended is
# MinMaxScaler.
# referece: https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.StandardScaler.html
# https://scikit-learn.org/stable/modules/preprocessing.html
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
X_train = sc.fit_transform(X_train) # Scaling to the range [0,1]
X_test = sc.fit_transform(X_test)X_trainarray([[-0.19511653, 0.42713604, -0.36510181, ..., -0.66659951,
-0.14895839, -0.31810766],
[-0.19511653, -2.34117451, 2.73896205, ..., -0.66659951,
-0.32815058, -0.31810766],
[-0.19511653, 0.42713604, -0.36510181, ..., 1.51653833,
-0.32815058, -0.31810766],
...,
[-0.19511653, 0.42713604, -0.36510181, ..., -0.66659951,
-0.29231215, -0.31810766],
[-0.19511653, 0.42713604, -0.36510181, ..., 1.51653833,
-0.32815058, -0.31810766],
[-0.19511653, 0.42713604, -0.36510181, ..., -0.66659951,
-0.32815058, -0.31810766]])y_trainarray([0, 0, 1, ..., 0, 1, 0])Part 2: Building FNN¶
########################################
# Part 2: Building FNN
#######################################
# Importing the Keras libraries and packages
import keras
from keras.models import Sequential
from keras.layers import Dense, InputInitializing the ANN¶
# Initialising the ANN
# Reference: https://machinelearningmastery.com/tutorial-first-neural-network-python-keras/
classifier = Sequential()
classifier<keras.engine.sequential.Sequential at 0x19cb8de02e0>Adding the input layer and the first hidden layer, 6 nodes, input_dim specifies the number of variables¶
# Adding the input layer and the first hidden layer, 6 nodes, input_dim specifies the number of variables
# rectified linear unit activation function relu, reference: https://machinelearningmastery.com/rectified-linear-activation-function-for-deep-learning-neural-networks/
# Adding the input layer with Input() and the first hidden layer
classifier.add(Input(shape=(len(X_train[0]),))) # Input layer
classifier.add(Dense(units=6, kernel_initializer='uniform', activation='relu'))
# Adding the second hidden layer
classifier.add(Dense(units=6, kernel_initializer='uniform', activation='relu'))
# Adding the output layer
classifier.add(Dense(units=1, kernel_initializer='uniform', activation='sigmoid'))Compiling the ANN¶
# Compiling the ANN,
# Gradient descent algorithm “adam“, Reference: https://machinelearningmastery.com/adam-optimization-algorithm-for-deep-learning/
# This loss is for a binary classification problems and is defined in Keras as “binary_crossentropy“, Reference: https://machinelearningmastery.com/how-to-choose-loss-functions-when-training-deep-learning-neural-networks/
classifier.compile(
optimizer = 'adam',
loss = 'binary_crossentropy',
metrics = ['accuracy']
)Fitting the ANN to the Training set¶
# Fitting the ANN to the Training set
# Train the model so that it learns a good (or good enough) mapping of rows of input data to the output classification.
# add verbose=0 to turn off the progress report during the training
# To run the whole training dataset as one Batch, assign batch size: BatchSize=X_train.shape[0#]
#classifierHistory = classifier.fit(X_train, y_train, batch_size = BatchSize, epochs = NumEpoch)Epoch 1/10
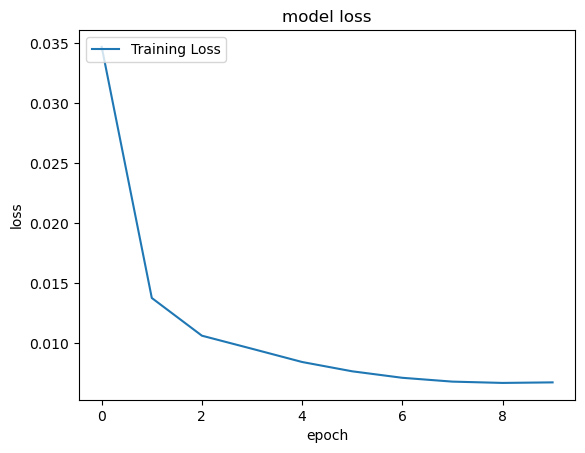
11333/11333 [==============================] - 26s 2ms/step - loss: 0.0347 - accuracy: 0.9885
Epoch 2/10
11333/11333 [==============================] - 26s 2ms/step - loss: 0.0137 - accuracy: 0.9960
Epoch 3/10
11333/11333 [==============================] - 25s 2ms/step - loss: 0.0106 - accuracy: 0.9969
Epoch 4/10
11333/11333 [==============================] - 25s 2ms/step - loss: 0.0095 - accuracy: 0.9974
Epoch 5/10
11333/11333 [==============================] - 26s 2ms/step - loss: 0.0084 - accuracy: 0.9977
Epoch 6/10
11333/11333 [==============================] - 26s 2ms/step - loss: 0.0076 - accuracy: 0.9979
Epoch 7/10
11333/11333 [==============================] - 26s 2ms/step - loss: 0.0071 - accuracy: 0.9981
Epoch 8/10
11333/11333 [==============================] - 25s 2ms/step - loss: 0.0068 - accuracy: 0.9981
Epoch 9/10
11333/11333 [==============================] - 25s 2ms/step - loss: 0.0067 - accuracy: 0.9981
Epoch 10/10
11333/11333 [==============================] - 26s 2ms/step - loss: 0.0067 - accuracy: 0.9982
Save the Fitted Model¶
classifier.save("fitted_FNN_model_SA.keras") # Save the model in HDF5 format
# Save the history
#import json
#with open("fitted_FNN_model_history_SA.json", "w") as f:
# json.dump(classifierHistory.history, f)Load the Model if Necessary¶
#from keras.models import load_model
#classifier = load_model("fitted_FNN_model_SA.keras") # Load the saved model
# Load the history
#import json
#with open("fitted_FNN_model_history_SA.json", "r") as f:
# classifierHistory = json.load(f)classifier.summary()Model: "sequential"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
dense (Dense) (None, 6) 702
_________________________________________________________________
dense_1 (Dense) (None, 6) 42
_________________________________________________________________
dense_2 (Dense) (None, 1) 7
=================================================================
Total params: 751
Trainable params: 751
Non-trainable params: 0
_________________________________________________________________
Evaluate the keras model for the provided model and dataset¶
from sklearn.metrics import classification_reportTraining Loss and Accuracy¶
loss_train, accuracy_train = classifier.evaluate(X_train, y_train)3542/3542 [==============================] - 4s 1ms/step - loss: 0.0061 - accuracy: 0.9984
print('Print the training loss and the accuracy of the model on the dataset')
print('Loss [0,1]: {0:0.4f} Accuracy [0,1]: {1:0.4f}'.format(loss_train, accuracy_train))Print the training loss and the accuracy of the model on the dataset
Loss [0,1]: 0.0061 Accuracy [0,1]: 0.9984
Testing Loss and Accuracy¶
loss_test, accuracy_test = classifier.evaluate(X_test, y_test)470/470 [==============================] - 1s 1ms/step - loss: 2.0659 - accuracy: 0.7774
print('Print the testing loss and the accuracy of the model on the dataset')
print('Loss [0,1]: {0:0.4f} Accuracy [0,1]: {1:0.4f}'.format(loss_test, accuracy_test))Print the testing loss and the accuracy of the model on the dataset
Loss [0,1]: 2.0659 Accuracy [0,1]: 0.7774
Part 3 - Making predictions and evaluating the model¶
########################################
# Part 3 - Making predictions and evaluating the model
#######################################
# Predicting the Test set results
y_pred = classifier.predict(X_test)
y_pred = (y_pred > 0.9) # y_pred is 0 if less than 0.9 or equal to 0.9, y_pred is 1 if it is greater than 0.9y_pred.shape(15017, 1)y_pred[:5]array([[False],
[False],
[ True],
[False],
[False]])y_test[:5]array([0, 1, 1, 0, 0])# summarize the first 5 cases
for i in range(10):
print('{} (expected {})'.format(y_pred[i], y_test[i]))[False] (expected 0)
[False] (expected 1)
[ True] (expected 1)
[False] (expected 0)
[False] (expected 0)
[False] (expected 1)
[False] (expected 0)
[False] (expected 1)
[ True] (expected 1)
[False] (expected 0)
Making the Confusion Matrix¶
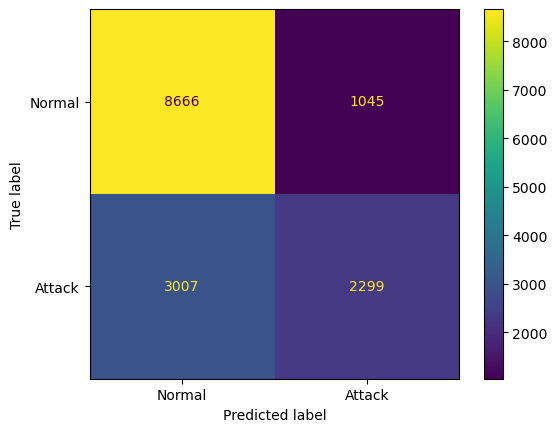
# Making the Confusion Matrix
# [TN, FP ]
# [FN, TP ]
from sklearn.metrics import confusion_matrix, ConfusionMatrixDisplay
cm = confusion_matrix(y_test, y_pred)
print('Print the Confusion Matrix:')
print('[ TN, FP ]')
print('[ FN, TP ]=')
print(cm)
print(classification_report(y_test, y_pred))
disp = ConfusionMatrixDisplay(confusion_matrix=cm, display_labels=['Normal', 'Attack'])
disp.plot()
plt.savefig('confusion_matrix_SA.png')
plt.show()Print the Confusion Matrix:
[ TN, FP ]
[ FN, TP ]=
[[8666 1045]
[3007 2299]]
precision recall f1-score support
0 0.74 0.89 0.81 9711
1 0.69 0.43 0.53 5306
accuracy 0.73 15017
macro avg 0.71 0.66 0.67 15017
weighted avg 0.72 0.73 0.71 15017
Part 4 - Visualizing¶
Receiver Operating Characteristic (ROC) Curve and Area Under Curve (AUC)¶
from sklearn.metrics import roc_curve, auc
import matplotlib.pyplot as plt
# Generate probabilities
y_pred_prob = classifier.predict(X_test).ravel()
# Calculate ROC curve
fpr, tpr, thresholds = roc_curve(y_test, y_pred_prob)
roc_auc = auc(fpr, tpr)
# Plot ROC curve
plt.figure()
plt.plot(fpr, tpr, label=f'ROC Curve (AUC = {roc_auc:.2f})')
plt.plot([0, 1], [0, 1], 'k--') # Random guess line
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Receiver Operating Characteristic')
plt.legend(loc='lower right')
plt.savefig('ROC_SA.png')
plt.show()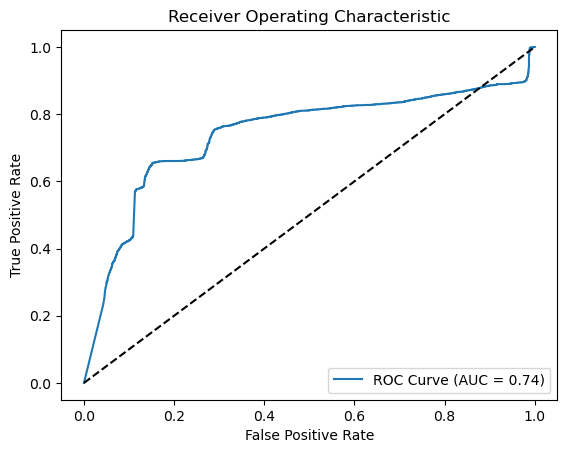
Plot the accuracy¶
########################################
# Part 4 - Visualizing
#######################################
# Import matplot lib libraries for plotting the figures.
import matplotlib.pyplot as plt
# You can plot the accuracy
print('Plot the accuracy')
# Keras 2.2.4 recognizes 'acc' and 2.3.1 recognizes 'accuracy'
# use the command python -c 'import keras; print(keras.__version__)' on MAC or Linux to check Keras' version
plt.plot(classifierHistory.history['accuracy'], label='Training Accuracy')
#plt.plot(classifierHistory['accuracy'], label='Training Accuracy')
plt.title('model accuracy')
plt.ylabel('accuracy')
plt.xlabel('epoch')
plt.legend(loc='upper left')
plt.savefig('accuracy_sample_SA.png')
plt.show()Plot the accuracy
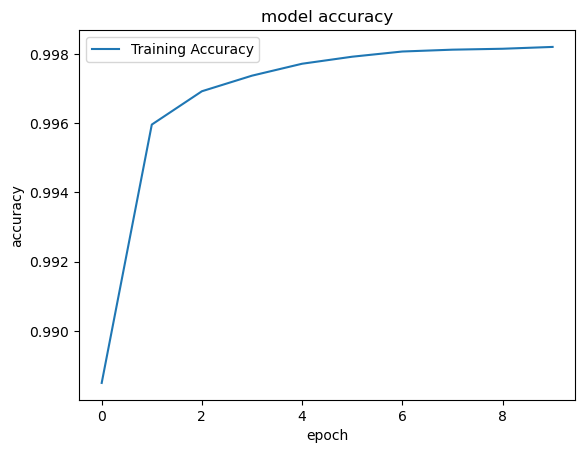
Plot the Loss¶
# You can plot history for loss
print('Plot the loss')
plt.plot(classifierHistory.history['loss'], label='Training Loss')
#plt.plot(classifierHistory['loss'], label='Training Loss')
plt.title('model loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(loc='upper left')
plt.savefig('loss_sample_SA.png')
plt.show()Plot the loss